Welcome to PathPPI
Version 1.0 (2013.11.16)
Ø Brief Introduction
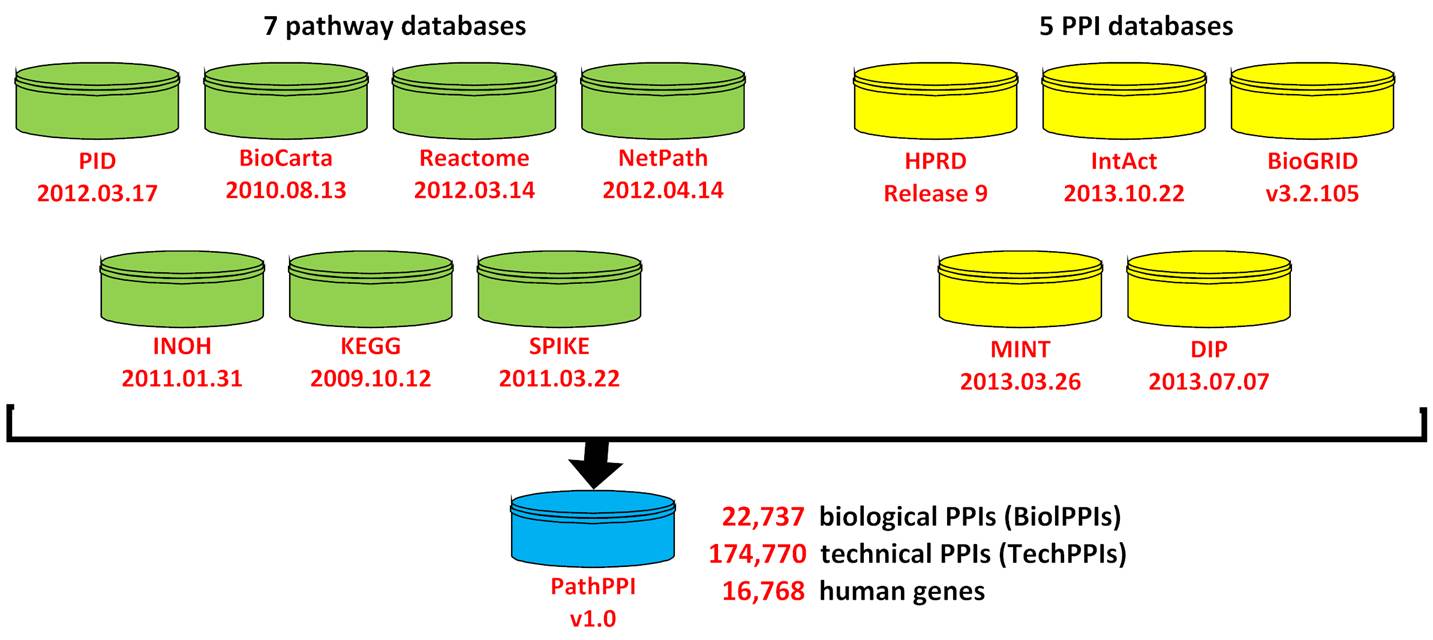
Integration of pathway and protein-protein interaction (PPI) data
can provide more information that could lead to new biological insights. PPIs
are usually represented by a simple binary model, whereas pathways are
represented by more complicated models. We developed a series of rules for
transforming protein interactions from pathway to binary model, and the protein
interactions from seven pathway databases, including PID, BioCarta, Reactome, NetPath, INOH, SPIKE and KEGG, were transformed based on these rules.
These pathway-derived binary protein interactions were integrated with PPIs
from other five PPI databases (HPRD, IntAct, BioGRID, MINT and DIP) to develop integrated dataset (named PathPPI). More information on protein interactions can be
preserved in PathPPI than other existing datasets.
Ø Categorisation of PathPPIs
|
PathPPI |
*Effect |
**Modification |
Directionality |
|
|
BiolPPI |
BiochemicalReactionRegulation (BRR) |
● |
● |
Directed |
|
TransportRegulation (TR) |
● |
|
Directed |
|
|
TransportWithBiochemicalReactionRegulation (TBRR) |
● |
● |
Directed |
|
|
ComplexAssemblyRegulation (CAR) |
● |
|
Directed |
|
|
ExpressionRegulation (ER) |
● |
|
Directed |
|
|
ComplexAssemblyInteraction (CAI) |
|
|
Undirected |
|
|
TechPPI |
GeneticInteraction (GI) |
|
|
Undirected |
|
MolecularInteraction (MI) |
|
|
Undirected |
|
*With three status: Activation (A),
Inhibition (I)
and Unspecified (U). **With 22 pairs of modification (see below) currently.
Ø 22 types of modifications in our current categorization of PathPPI
|
Modification |
Abbreviation
in PathPPI |
|
Acetylation/Deacetylation |
Ac+/Ac- |
|
Biotinylation/Debiotinylation |
Biotin+/Biotin- |
|
Decanoylation/Dedecanoylation |
Decanoyl+/Decanoyl- |
|
Dimethylation/Dedimethylation |
Me2+/Me2- |
|
Farnesylation/Defarnesylation |
Farnesyl+/Farnesyl- |
|
Fucosylation/Defucosylation |
Fucosyl+/Fucosyl- |
|
Galactosylation/Degalactosylation |
Gal+/Gal- |
|
Geranylgeranylation/Degeranylgeranylation |
GG+/GG- |
|
Glucosylation/Deglucosylation |
Glucosyl+/Glucosyl- |
|
Glycosylation/Deglycosylation |
Glycosyl+/Glycosyl- |
|
Glycylation/Deglycylation |
Glycyl+/Glycyl- |
|
Hydroxylation/Dehydroxylation |
Hydroxyl+/Hydroxyl- |
|
Lipoylation/Delipoylation |
Lipoyl+/Lipoyl- |
|
Methylation/Demethylation |
Me+/Me- |
|
Myristoylation/Demyristoylation |
Myristoyl+/Myristoyl- |
|
Octanoylation/Deoctanoylation |
Octanoyl+/Octanoyl- |
|
Palmitoylation/Depalmitoylation |
Palmitoyl+/Palmitoyl- |
|
Phosphopantetheine/Dephosphopantetheine |
Phosphopantetheine+/Phosphopantetheine- |
|
Phosphorylation/Dephosphorylation |
Phos+/Phos- |
|
Sumoylation/Desumoylation |
Sumo+/Sumo- |
|
Trimethylation/Detrimethylation |
Me3+/Me3- |
|
Ubiquitination/Deubiquitination |
Ub+/Ub- |
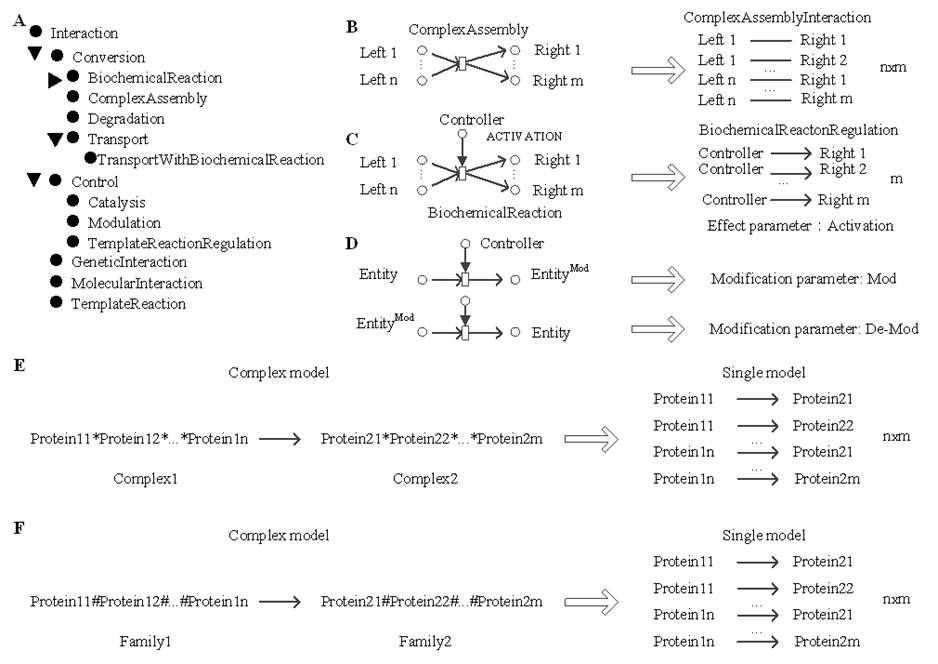
Ø
The
transformation from BioPAX to PathPPI
model
|
|
1.
BioPAX Level 3 contains five types of molecular
interactions. Control and Conversion have subclasses. 2.
For
ComplexAssembly, we specified that each input entity
have a CAI with each output entity in the PathPPI
model. 3.
For
Control, we defined five types of interactions between the controller and
products of controlled interaction for each controlled Conversion
interaction. For example, if the controlled interaction is BiochemicalReaction, the controller has a BiochemicalReactionRegulation interaction with each
product. Effect parameters can be obtained from controlType. 4.
Modification
parameters can be obtained by comparing the modification state of the entity before
and after reaction. (E,
F) Complex model allows PathPPI entities to be
complexes or families, in contrast to a single model where each protein from
one entity in the complex model interacts with each protein from another
entity. |
Ø BiolPPI composition
of the 7 BiolPPI databases
|
Ø Proportions
of different types of entity in the 7 pathway databases
The ratios
of different types of entities occupied all entities were shown in brackets. (Complex model) |
Ø
BiolPPI composition
of the 7 BiolPPI databases (Complex model)
|
Dataset |
BRR |
TR |
TBRR |
CAR |
ER |
CAI |
|
PID |
2,160 |
200 |
131 |
709 |
1,700 |
5,195 |
|
BioCarta |
1,149 |
46 |
1 |
256 |
242 |
1,143 |
|
Reactome |
1,519 |
0 |
0 |
0 |
237 |
0 |
|
NetPath |
582 |
0 |
0 |
0 |
0 |
0 |
|
INOH |
198 |
2 |
0 |
1 |
0 |
691 |
|
KEGG |
1,823 |
0 |
0 |
0 |
284 |
0 |
|
SPIKE |
4,504 |
0 |
0 |
0 |
1,776 |
0 |
|
BiolPPI |
10,627 |
239 |
132 |
948 |
3,938 |
6,853 |
Ø
Download
|
PathPPI v1.0 (197,507 PPIs) |
BiolPPI v1.0 (22,737 PPIs) TXT |
|
TechPPI v1.0 (174,770 PPIs) TXT |
|
===================================================================================================================
PathPPI was developed by the
Lab of Systems Biology, Institutes of Biomedical Sciences, Fudan University.
This work is supported by: MOST-863/S973 projects [2012AA020201,
2013CB910802, 2010CB912700], National Natural Science Foundation Projects
[31000379, 31000587, 31000591],
and Chinese State Key Project Specialized for Infectious Diseases
[2012ZX10002012-006].
Contact to: 051022003@fudan.edu.cn